Documentation for ixdat¶
The in-situ experimental data tool¶
With ixdat, you can import, combine, and export complex experimental datasets
as simply as:
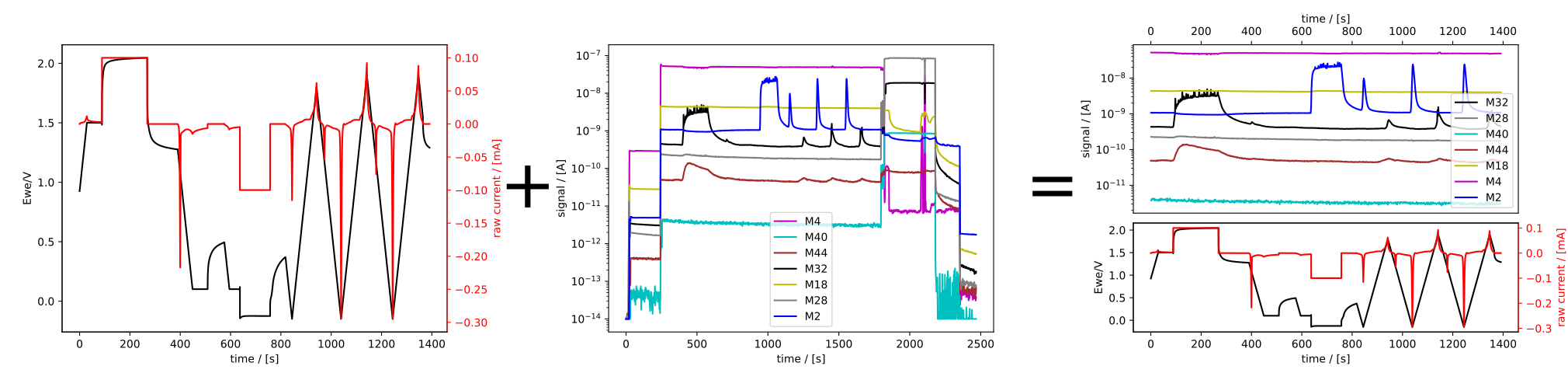
ec = Measurement.read_set("awesome_EC_data", reader="biologic")
ec.plot_measurement()
ms = Measurement.read("2021-03-30 16_59_35 MS data.tsv", reader="zilien")
ms.plot_measurement()
ecms = ec + ms
ecms.plot_measurement()
ecms.export("my_combined_data.csv")
Output:
Or rather, than exporting, you can take advantage of ixdat’s powerful analysis
tools and database backends to be a one-stop tool from messy raw data to public
repository accompanying your breakthrough publication and advancing our field.
The documentation¶
Welcome to the ixdat documentation. We hope that you can find what you are looking for here!
The Introduction has a list of the techniques and file types supported so far.
This documentation, like ixdat itself, is a work in progress and we appreciate any
feedback or requests here.
Note, we are currently compiling from the [user_ready] branch, not the master branch.